Laurent Jacob
Researcher in machine learning and statistics for genomics
I am a senior CNRS researcher located at the LCQB lab of Sorbonne Université in Paris.
I work on machine learning and statistics to answer questions in genomics. I am particularly interested in microbial genomics, notably to better understand and predict drug resistances, and in evolutionary genomics. Part of my work also aim at better understanding neural networks for biological sequences. See the Projects section for some examples.
I received an MSc in machine learning from ENS Cachan (now ENS Paris-Saclay) in 2006, and a Ph.D. from Mines ParisTech in 2009, where I worked on regularized machine learning for computational biology. Between 2010 and 2012 I was a postdoctoral fellow at the statistics department of UC Berkeley. I joined the CNRS in 2013 and worked at the LBBE lab in Université Lyon 1 until 2023.
You can reach me at firstname (dot) lastname (at) cnrs (dot) fr.
Projects

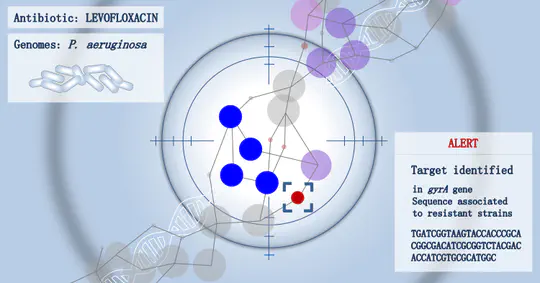
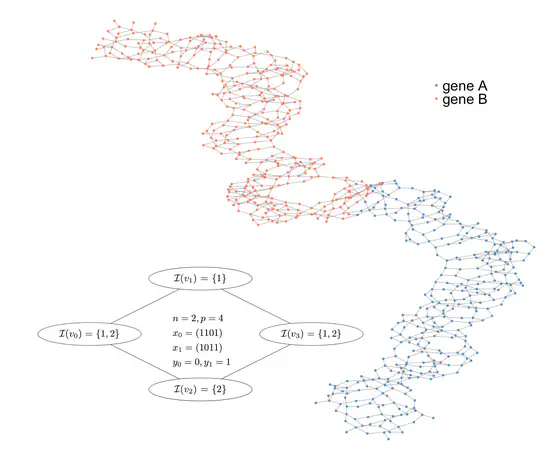
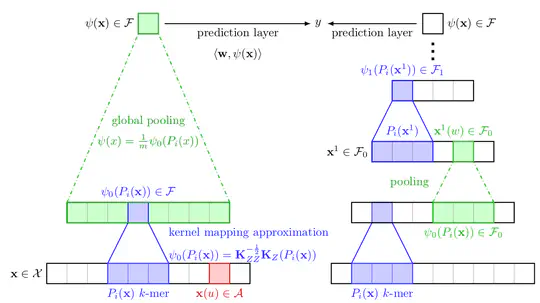
Some recent publications
Community
I was recently involved in the organization of several scientific events:
- LEGEND 2025, a conference on machine learning for evolutionary genomics.
- I am co-chairing the Sequence Analysis track of the 2025 ISMB conference.
- LEGEND 2024, a conference on machine learning for evolutionary genomics.
- LEGO, a French working group on machine learning for genomics. We organize national meetings to gather researchers interested in this topic.
- MLMG 2022, an ECML workshop on machine learning for microbial genomics.
- I co-chaired the Systems Biology and Networks track of the 2021 ISMB conference.
- The 2020 prospective meeting on data science, AI and biology from the biology and computer science institutes of CNRS.